MATERIALS AND METHODS:
In the current experiment, males from the Dr/Tm3Ser Sb stock were crossed to virgin females from the stock containing CyO balancer chromosome on Dahomey background (see step1 in figure 8). In the next generation, males bearing the Dr and CyO traits (CyO; Dr) were selected. Hundred CyO; Dr males were mutagenized using 10% EMS in a sugar solution. The males were removed away from the food for approximately 24 hours before treatment. In addition, keeping them under the fume hood for approximately 3 hours before treatment further dehydrated them. The deprival from food and moisture was meant to ensure that they were hungry and thirsty when offered the mutagen and ingested it (step2). The mutagenized males were then mated with virgin Dahomey females (step2).
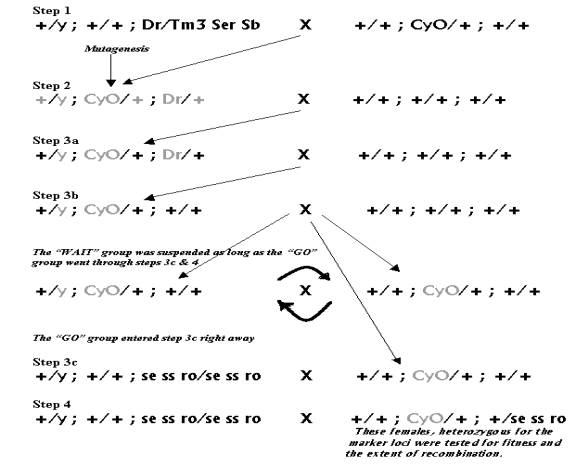
Figure 8: The above series of crosses was undertaken to get female flies carrying mutagenized CyO chromosome isolated on the wildtype background as well as the recessive markers on the third chromosome, used to record the extent of recombination.
In the next generation (step3) of the Step2 cross, 90 CyO; Dr males were chosen in batches of 15, such that all the males from one batch had emerged on the same day. Thus there was a range of variation among the males based on the length of developmental period. This variation in the length of development, among batches of males that emerged over the six-day collection period, was theorized to reflect a range of variation in the fitness of the chosen males. The males that were first to emerge were expected to have the greatest fitness, while the males that emerged on the sixth day were expected to have the lowest fitness. Furthermore, since EMS does not uniformly mutate all the cells it affects, therefore every sperm cell in a step2 male was expected to be mutated differently and thus each one of them was unique with respect to the mutations it acquired. Hence each of the 90 males chosen for step3a should have had a different CyO chromosome, with its unique set of EMS-induced mutations. While each of the step3a males was unique with respect to the CyO chromosome it bore, it had half of the mutagenized genome of its father replaced by the non-mutagenized genome from its mother. Hence among the 3a males, the non-mutagenized portion of the genome was theorized to be genetically similar, because all the mothers came from the same lab population.
The 3a males were again crossed to virgin Dahomey females and from their progeny, males with only the CyO trait were selected. The 3b males had inherited only two out of the three mutagenized chromosomes that their fathers bore, that is the “y” and the “CyO” (see figure 8, step3b). Like the two generations before them, the 3b males were also mated to virgin Dahomey females. The progeny of 3b males was split into two groups: the first group was meant to continue the lines with mutagenized CyO in them, while the second group went through step 3c. To continue the lines, CyO males from the progeny of 3b males were mated to their CyO sisters. In addition, five CyO females from each of the surviving 78 lines were mated to sets of four males from the stock containing se ss ro recessive markers on the third chromosome; this was step 3c of the protocol. The third chromosome markers mentioned earlier were introduced into the genomes of the experimental flies in order to record the recombination events that take place between them.
Heterozygous females from the progeny of the 3c cross were backcrossed in step 4 to the se ss ro males. This three-point cross (comprising steps 3c & 4) was meant to express the extent of meiotic recombination in the step4 mothers, as the proportion of their offspring bearing recombinant phenotypes. One thing to note here is that over the series of earlier crosses, we got our experimental flies rid off all the mutagenized chromosomes except the CyO balancer. Therefore it was theorized that any significant difference in fitness among the step 4 females, if observed, would be due to the effect of the mutagenized CyO balancer, since the rest of the genome of all the experimental lines was derived from related stocks under similar laboratory conditions (see figure 8). In our experiment, we set the female fecundity as a measure of fitness, which was reflected in the number of viable offspring that a female produced.
Thus in the step 4 of the protocol (figure 8), 3 sets of 3 virgin females were collected from each of the 39 experimental lines, to a total of 117 sets. The females, at the age of approximately two days were mated to se ss ro males (4♂s to3♀s), in vials containing food. The parents were removed from the culture medium roughly 72+1 hours after mating. The vials were scored for the number of parental (two phenotypic classes) and recombinant (six phenotypic classes) offspring, fifteen days after mating.
The extent of recombination was determined using the following formulas:
Amount of recombination between se and ss:
r1 = (number of se-ss recombinants + number of double recombinants)
(total number of offspring in the vial)
Amount of recombination between ss and ro:
r2 = (number
of ss-ro recombinants + number of double recombinants)
(total number of offspring in the vial)
Net amount of crossing over:
R = (number of se-ss
recombinants + ss-ro recombinants + 2*double recombinants)
(total number of offspring in the vial)
or
R = r1 + r2.
However the eight phenotypic classes were not all equally viable and the numbers of offspring in the reciprocal phenotypic classes (+++ & se ss ro, se ss + & ++ ro, se ++ & + ss ro, etc) were widely different from each other. Therefore the errors due to differential viability, in the calculation of the amount of recombination were corrected using the method adopted by A.G. Clark, in his 1981 publication on the estimation of linkage. Nonetheless, the data points on the graph of fitness vs. recombination looked qualitatively very similar to what they were before applying Clark’s correction (see results section). Therefore this report will only discuss the estimates of recombination that employed the classical approach of calculation, and not the ones taking viability into account, since both methods reveal a similar relation between fitness and recombination.