
|
In collaborative studies with proteomic experts Dr. Thomas Kislinger (Ontario Cancer Institute, U of Toronto) and Dr. Andrew Emili (Donnelly Centre for Cellular and Biomolecular Research, U of Toronto) we have been developing methodologies for the large scale study of proteomic expression profiles in cardiovascular diseases. Together with Dr Peter Liu (University of Ottawa Heart Institute) we have been applying these techniques to study the mechanisms involved in the progression of heart failure in mouse models, as well as in human patients. These experiments are undertaken with the aim of uncovering new cellular pathways that are involved in the disease in order to develop new therapeutic targets. As well, we have been attempting to identify new blood-borne biomarkers of cardiovascular disease to develop new screening tools.
|

|
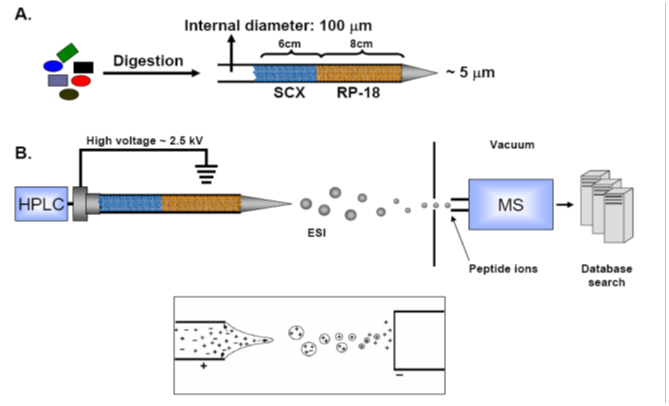
Shown above is the gel-free, shotgun mass spectrometry, expression system employed to identify proteins in complex mixtures. |
|
Active Research Projects |
|
The overall objectives of this project are to provide a clearer understanding of the role of phospholamban (PLN) in the normal working of the heart, and what are the cellular mechanisms in human PLN genetic diseases that result in dilated cardiomyopathy. Further studies of the formation of the PLN–SERCA2a complex might provide further clues as to the mechanisms that underlie PLN regulation and provide new insights into ways to design chemicals that would disrupt this interaction selectively. Our aim is to provide such novel approaches to aid in the diagnosis and treatment of cardiac diseases resulting from defects in the regulation of intracellular calcium concentrations.
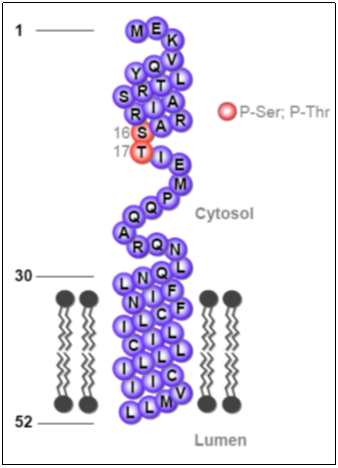
Shown, at left, is a schematic diagram of the 52 amino acids that constitute PLN, together with the phosphorylation sites of PLN (Ser-16 and Thr-17)
These experiments have been performed together with several international collaborative initiatives involving Dr. Litsa Kranias (U Cincinnati) , Dr Peter Backx (U Toronto), Dr. Jon Seidman (Harvard), Dr. Christine Seidman (Harvard), and Dr. David MacLennan (U Toronto). |
|
2. Phospholamban (PLN) in Cardiac Muscle |
|
1. Applied Proteomics in Cardiac Muscle Diseases |
|
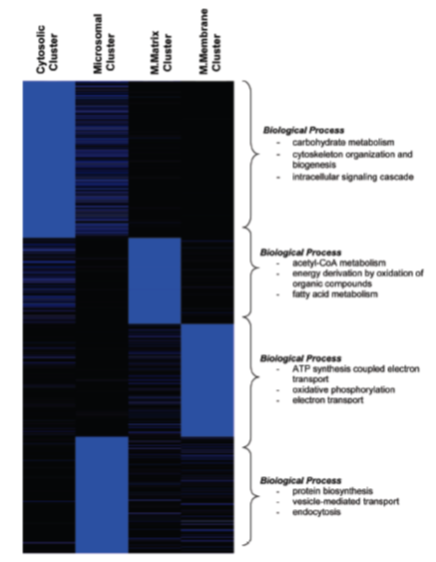
Hierarchical clustering of cardiac proteins with the major sub-cellular compartments. Shown is an unbiased self-organizing heat map demonstrating the segregation of proteins into 4 main clusters representing the 4 sub-cellular fractions. To the right are key “Biological Process” GO terms found to be significantly enriched in each respective cluster. |
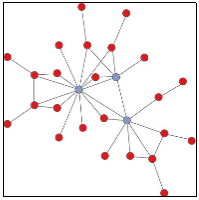
As we complete our large scale proteomic investigations of mouse and human models of heart failure, we are extending our findings by developing advanced protein-protein interaction (PPI) map of protein networks affected in heart failure. We are in the process of using this information to identify novel cellular targets for development of therapeutic interventions.Our specific interest and expertise in this project is the application of phenotypic analysis, cellular imaging and protein–protein interaction analyses to identify key protein interactions. We do this by using mammalian cell lines, transgenic and knockout mice, primary cardiac muscle-derived cell cultures, and patient-derived cardiac tissues, to understand cardiac function and disease. We use a combination of molecular biology with computational analyses, where the protein interaction data will be integrated into existing and newly developed bioinformatic tools being maintained within the University of Toronto for detailed analyses and sharing. Our initial focus is the identification of protein interactions involving several critical calcium regulatory proteins; however, we are extending our abilities rapidly to other key proteins found to be important in heart failure. The emphasis will be on discovering novel protein interactions critical for understanding the progress of cardiomyopathy. The ultimate aim of this project is to provide clinically relevant therapeutic methods that can be applied to the diagnosis and treatment of heart failure in human patients. |
|
University of Toronto, Toronto, Ontario, M5G 1L6 |