Part 0: Introduction
Last Meeting Summary
- The needed parts have been requested, Randy has said he’ll ship on Nov 11th. Enzymes and buffers have been obtained.
- The temperature sensor project name is tentatively the “Cell-see-us” Thermometer, thank Andrew for that one :)
- It’s been agreed that both the temperature sensor and etch-a-sketch will be assembled and tested. Note that both can be assembled in a total of 10 addition operations.
- It’s been generally decided that the undergrads (Hannah/Emanuel/Joseph) will be working together at Steve’s lab (MSB4232) to combine all the parts. Please let us know if you want in for scheduling purposes…
To Do
- Assemble, transform E. coli, do multiple trials. Make video (I’m hoping there’s a camcorder available?).
- I can make the presentation, everyone please email me any materials/thoughts/suggestions you think may be helpful.
Part 1: Overview of Cell-See-Us Thermometer and Bacterial Etch-a-Sketch
Tuneable Temperature Sensor v2 (using LacI ts + other parts from Registry 7.05) [BBa_J11021]
http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=6155
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
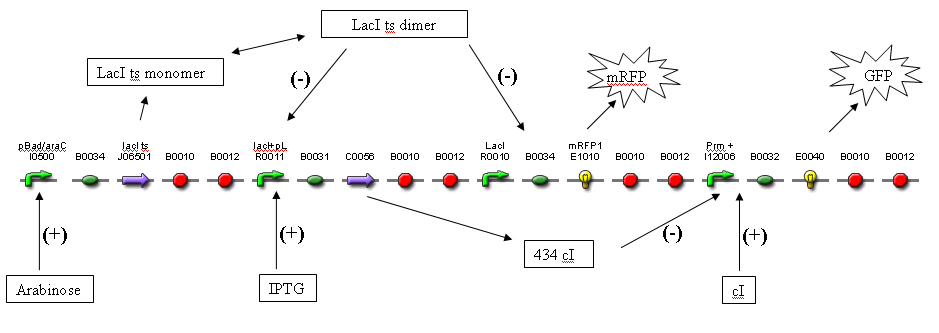
Explanation Summary:
Keeping the cells in a L-arabinose solution induces the cells to produce a specific steady state value of lacI monomers. The portion of these monomers that is in dimer form decreases with temperature. As temperature increases, the LacI dimer concentration decreases, resulting in reduced inhibition of the R0010 promoters. Hence RFP concentration increases. At the same time, more cI is produced, which inhibits Prm +, resulting in less GFP at steady state.
Explanation Details:
1) Consider a system with just the “LacI” (pLac) promoter and associated coding regions. The cells are in an IPTG solution, which results in the promoters being induced, resulting in 434 cI and RFP being produced at a “maximal” rate.
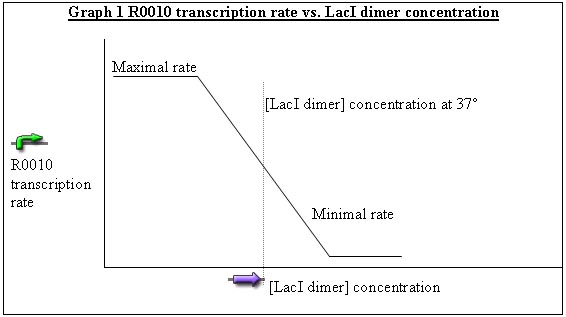
2) Now consider some LacI ts added to the system, which forms a dimer at some equilibrium concentration. At [LacI dimer] = 0, 434 cI and RFP are produced at maximal rates. At some concentration of LacI dimer, the transcription rate becomes “minimal” since LacI inhibits the “LacI” promoter (pLac). Between the maximal and minimal rates, there is a linear region where transcription rate gradually decreases as a function of LacI concentration. (see Graph 1)
3) The pBad promoter can be induced by keeping the cells in a solution of L-arabinose. When induced, lacI ts is produced. The concentration of L-arabinose can be set so that at 37˚ the [LacI dimer] concentration is centered in the linear region. Hence as temperature deviates from 37˚, R0010 changes accordingly.
4) As the R0010 transcription rate changes, the RFP concentration steady state value changes accordingly. Also, the cI steady state concentration changes.
5) To increase the visual contrast, Prm + driving GFP has been added to the system. For example, if the temperature increases from 37˚ to 42˚, the LacI dimers disassociate more readily, and [LacI dimer] decreases. R0010 transcription rate increases, resulting in higher [RFP] and [434 cI] steady state values. Since 434 cI represses Prm, the Prm transcription rate will decreases, resulting in a lower [GFP]. It is hoped that the decrease in steady state [434 cI] will indeed result in a noticeable change in [GFP], this depends on [434 cI] at 37˚ being in the linear region of a Prm + transcription rate vs. [434 cI] graph. (A way of tuning this will likely be needed – investigating currently)
Tuneable Temperature Sensor v3 (Registry 7.05 DNA-1 + Harvard Parts) [BBa_J11022]
http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=6160
|
pBad/araC |
|
lacI
ts |
|
|
lacI+pL |
|
|
|
|
Prm
+ |
|
|
|
|
LacI |
|
mRFP1 |
|
|
|
|
|
Explanation Summary:
The same as BBa_J11022, however the parts have been moved around so that currently available parts can be used to reduce the overall number of parts (no change in functionality). Also, some minor substitutes have been made (ex. R0010->R0011) to further reduce the number of parts. For synthesis, only 8 parts have to be assembled together.
|
pBad/araC |
|
|
|
|
Prm
+ |
|
|
|
|
|
Bacterial Whiteboard by Hannah (w/ Minimal Available Parts) [BBa_J11031]
http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=6154
|
tetR |
|
lacZ a |
|
|
lacI+pL |
|
ECFP |
|
|
|
|
|
Explanation Summary:
When lactose is added to the system, bind to lac repressor (endogenous) therefore CFP (fast degrading) produced - glow cyan. When Tet added to the system, drive transcription of lacZ beta galactosidase which hydrolyses latose and lac repressor active. Stop Glowing cyan. ***NOTE*** This system will ONLY WORK IN LAC Z- bugs!!!!!
For synthesis, only 4 parts have to be assembled together:
|
tetR |
|
lacI+pL |
|
|
|
|
Part 2: Temperature Sensor Modeling
Note: “In the presence of lactose or IPTG, an analog of lactose, LacI is unable to correctly bind and inhibit transcription.” [http://parts2.mit.edu/r/parts/partsdb/view.cgi?part_id=185]
Part 3: Assembly Schedule
|
Part |
Day 1 |
Day 2 |
Day 3 |
|
I0500+ J06801 |
(I0500+ J06801)+( B0031+ C0056) |
{(I0500+ J06801)+( B0031+ C0056)}+{ (B0015+ I12006)+( E0240+ J04450)} |
|
|
B0031 |
B0031+ C0056 |
||
|
C0056 |
|||
|
B0015 |
B0015+ I12006 |
(B0015+ I12006)+( E0240+ J04450) |
|
|
I12006 |
|||
|
E0240 |
E0240+ J04450 |
||
|
J04450 |
|||
|
R0040 |
R0040+ E0433 |
(R0040+ E0433)+( R0011+ E0422)
|
|
|
E0433 |
|
||
|
R0011 |
R0011+ E0422 |
|
|
|
E0422 |
|
BBa_J11022
|
pBad/araC |
|
|
|
|
Prm
+ |
|
|
|
|
|
BBa_J11031
|
tetR |
|
lacI+pL |
|
|
|
|
Part 4: Part Locations
|
Part |
Registry 7.05 DNA-1 Well |
|||||||||||||||||||||||||||||||||||
|
I0500 |
|
|||||||||||||||||||||||||||||||||||
|
J06801 |
|
|||||||||||||||||||||||||||||||||||
|
B0031 |
3I |
|||||||||||||||||||||||||||||||||||
|
C0056 |
18L |
|||||||||||||||||||||||||||||||||||
|
B0015 |
1I |
|||||||||||||||||||||||||||||||||||
|
I12006 |
|
|||||||||||||||||||||||||||||||||||
|
E0240 |
16A |
|||||||||||||||||||||||||||||||||||
|
J04450 |
|
|||||||||||||||||||||||||||||||||||
|
R0040 |
7O |
|||||||||||||||||||||||||||||||||||
|
E0433 |
14K |
|||||||||||||||||||||||||||||||||||
|
R0011 |
7M |
|||||||||||||||||||||||||||||||||||
|
E0422 |
11G |
|||||||||||||||||||||||||||||||||||