Our interdisciplinary and innovative program incorporates six major themes, as described below.
1. Functional Genomics
2. Chemical Genomics
3. Mechanisms of Drug Resistance and Disease
4. Microbiome in Health and Disease
5. Structure-Guided Drug Design
6. Experimental Evolution
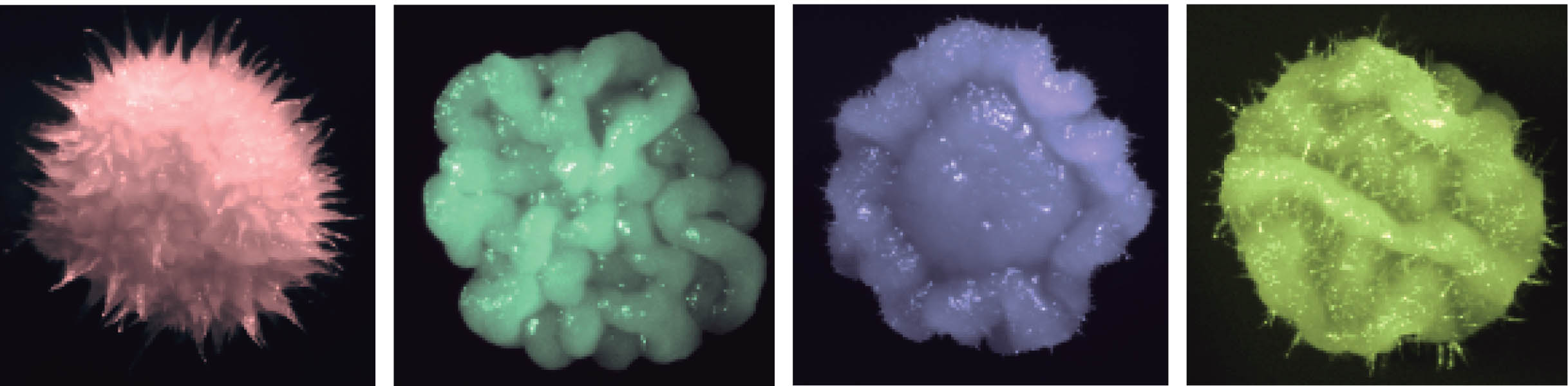
Functional Genomics
Functional genomics focuses on genome-scale exploration of gene function to understand biological systems.
Our functional genomics efforts are enabled by genome-scale mutant collections of Candida albicans, which is a leading fungal pathogen of humans. Candida species account for 88% of all hospital-acquired fungal infections and cost the health care system over $1 billion annually in the United States alone. C. albicans is the primary cause of systemic candidiasis with mortality rates of ~40%, even with current treatments.
We utilize functional genomics in many facets of our research, including global analysis of fungal drug resistance and morphogenesis, which is a key virulence trait. Mutants that are unable to transition between yeast and filamentous growth are typically attenuated in virulence. The current paradigm is that yeast cells are critical for colonization and dissemination, while filaments are required for tissue invasion and expression of virulence factors such as adhesins and proteases.
We have developed powerful barcode sequencing based pooled assays to complement our high-content imaging platform. Our work provides a global view of circuitry governing drug resistance, morphogenesis, and virulence, and identifies novel targets for antifungal drug development.
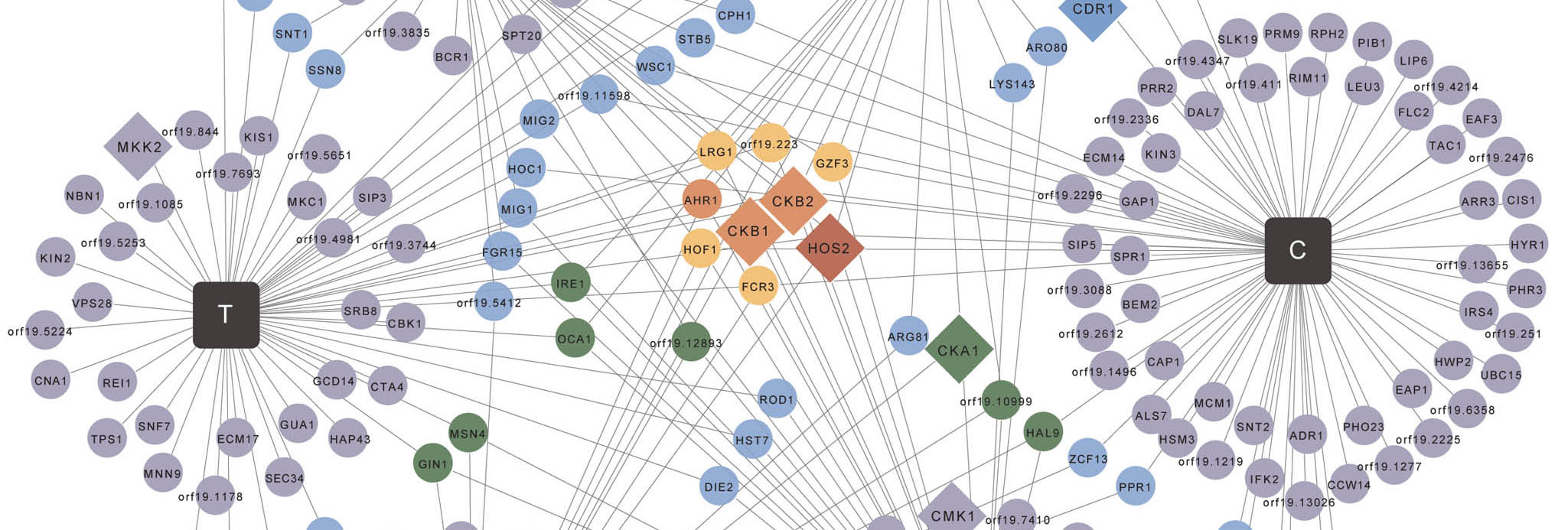
Chemical Genomics
Chemical genomics leverages small molecules to probe gene function and understand biological systems.
We use chemical genomics to map genetic interaction networks that control key virulence traits, and to link biologically active small molecules to their targets.
Small molecules provide a powerful approach to detect genetic interactions when a mutant allele of one gene causes an unexpected phenotype in the presence of a small molecule. Chemical genomics is crucial to mapping genetic interactions in organisms for which classical genetic approaches are hampered by the lack of a complete sexual cycle, as with C. albicans. Chemical genomics also provides a powerful strategy to identify the targets of small molecules based on the principle that reducing the dosage of a target will confer hypersensitivity to the cognate small molecule.
Our work reveals key principles by which connectivity in genetic networks can illuminate the genetic architecture of biological pathways, and identifies novel regulators of drug resistance and morphogenesis, as well as new therapeutic targets.
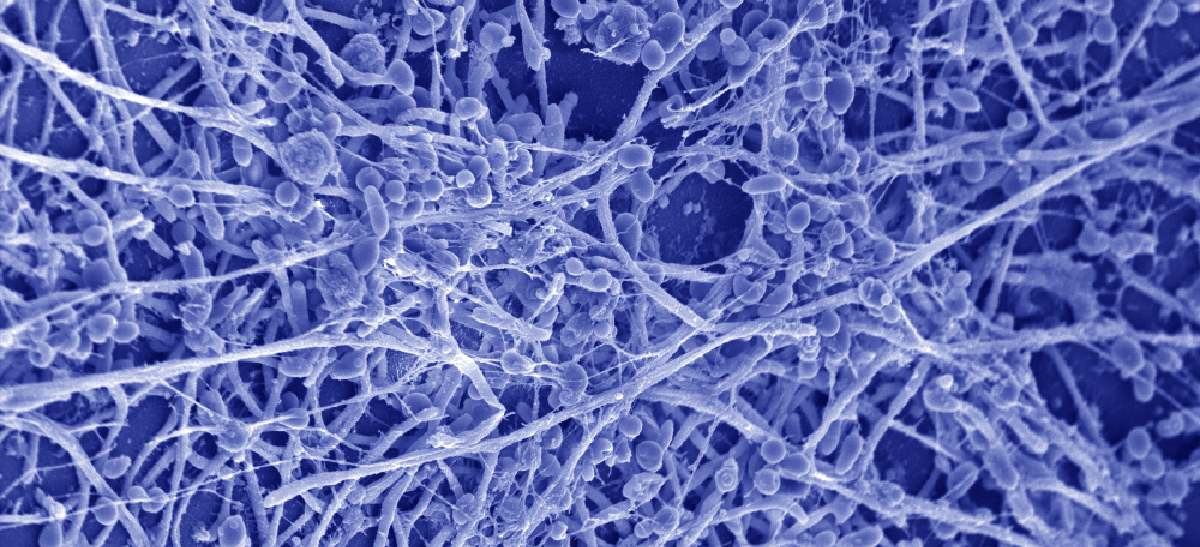
Mechanisms of Drug Resistance and Disease
Treatment of invasive fungal infections is notoriously difficult, with only three classes of antifungals for invasive fungal infections, compared to more than two dozen classes for bacterial infections. Resistance to each class of antifungal has emerged as a clinical problem.
We use detailed molecular genetic approaches to dissect the mechanisms that allow fungal pathogens to evade the drugs we use to kill them, and cause life-threatening infectious disease.
As just one example, our work reveals that the molecular chaperone Hsp90 is the Achilles heel of fungal pathogens, as it orchestrates the cellular circuitry required for stress response, drug resistance, morphogenesis, and virulence. Hsp90 enables drug resistance by stabilizing key regulators of cellular signaling such as the protein phosphatase calcineurin and mitogen activated protein kinase Mkc1. Hsp90 governs morphogenesis by repressing Ras1-PKA signaling and through more specialized cellular circuitry. Hsp90 function is subject to complex regulation by co-chaperones and by post-translational modification, including phosphorylation and acetylation.
We also leverage chemical biology approaches to screen for small molecules that reverse drug resistance, and to link the bioactive small molecules to their cellular target. Many small molecules with antifungal activity also modulate key virulence traits such as morphogenesis.
Our focus on fungal pathogenesis also extends to understanding the mechanisms by which fungi sense and respond to temperatures in the human host, form drug-resistant biofilms on catheters and indwelling medical devices, and interact with host immune cells.
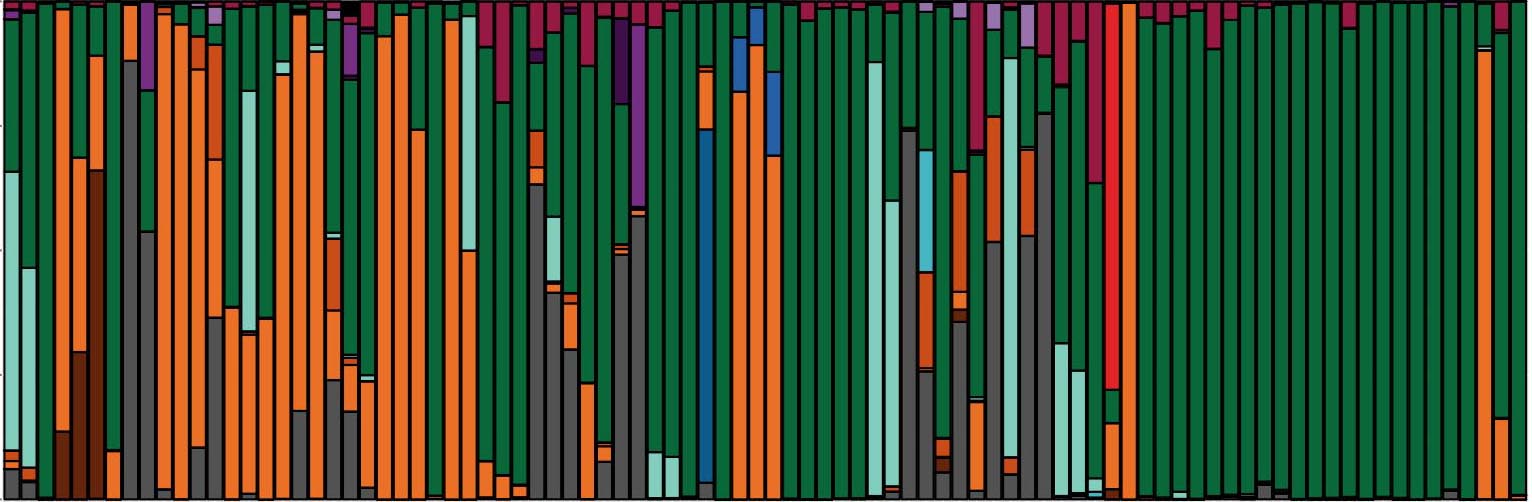
Microbiome in Health and Disease
The human body is home to a remarkable number of microbes, collectively referred to as the human microbiome, which have a profound impact on diverse facets of human biology and disease.
We have developed powerful approaches to characterize the fungal microbiome leveraging high-throughput sequencing of the ribosomal RNA internal transcribed spacer 1 (ITS1), coupled with phenotypic and genotypic analyses fungal isolates.
This clinical genomic approach has allowed us to track fungal communities in patient populations over the course of clinical exacerbation and treatment. This reveals that fungal communities found in the lung of patients with respiratory diseases are distinct from those in healthy people. It also reveals mechanisms of pathogen adaptation over the course of chronic infection.
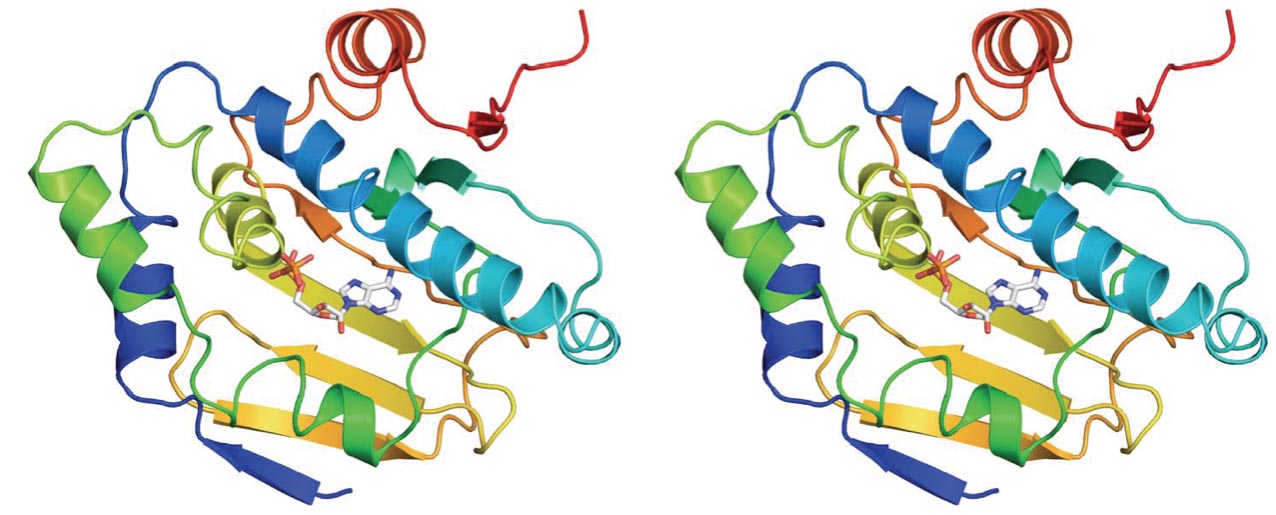
Structure-Guided Drug Design
Solving three-dimensional structures of drug targets allows for rational and precise drug discovery and development.
We established that targeting Hsp90 provides a powerful strategy to treat fungal infections. In species of Candida and Aspergillus, genetic depletion or pharmacological inhibition of Hsp90 abolishes resistance to the most widely deployed antifungals, including azoles, echinocandins, and polyenes. We have demonstrated that inhibiting Hsp90 function renders resistant fungal pathogens more responsive to treatment, impedes the evolution of drug resistance, and leads to clearance of otherwise lethal infections.
Hsp90 is an attractive target for drug development, and there has been phenomenal progress in the discovery of potent compounds that selectively inhibit this chaperone, driven in large part by anti-cancer activity of Hsp90 inhibitors. All Hsp90 inhibitors currently undergoing clinical development have been optimized to inhibit the human chaperone and exhibit relatively poor activity against fungi.
We have assembled an interdisciplinary team to solve the crystal structure of the N-terminal domain of C. albicans Hsp90. Guided by the key differences in the X-ray crystal structures of fungal and human Hsp90, we have been able for the first time to design, synthesize and biochemically characterize lead inhibitors of fungal Hsp90 with >10-fold selectivity relative to the human isoform.
Experimental Evolution
Experimental evolution focuses on testing hypotheses regarding evolution using controlled laboratory experiments.
Drug combinations are emerging as a promising strategy to treat fungal infections, but the mechanisms by which they evolve and the fitness consequences have remained largely enigmatic.
We have leveraged experimental evolution coupled with genome sequencing and high-resolution fitness measurements to map the genetic architecture of adaptation to antifungal drug combinations. We have complemented these studies with genome-scale analyses of how drug resistance evolves in the human host, and have identified fitness trade-offs in which the evolution of drug resistance is accompanied by hypersensitivity to killing by macrophages.
This work provides a foundation for predicting and preventing the evolution of drug resistance, and reveals fitness trade-offs that may minimize the evolution and persistence of resistance in pathogen populations.
1. Functional Genomics
2. Chemical Genomics
3. Mechanisms of Drug Resistance and Disease
4. Microbiome in Health and Disease
5. Structure-Guided Drug Design
6. Experimental Evolution

Functional Genomics
Functional genomics focuses on genome-scale exploration of gene function to understand biological systems.
Our functional genomics efforts are enabled by genome-scale mutant collections of Candida albicans, which is a leading fungal pathogen of humans. Candida species account for 88% of all hospital-acquired fungal infections and cost the health care system over $1 billion annually in the United States alone. C. albicans is the primary cause of systemic candidiasis with mortality rates of ~40%, even with current treatments.
We utilize functional genomics in many facets of our research, including global analysis of fungal drug resistance and morphogenesis, which is a key virulence trait. Mutants that are unable to transition between yeast and filamentous growth are typically attenuated in virulence. The current paradigm is that yeast cells are critical for colonization and dissemination, while filaments are required for tissue invasion and expression of virulence factors such as adhesins and proteases.
We have developed powerful barcode sequencing based pooled assays to complement our high-content imaging platform. Our work provides a global view of circuitry governing drug resistance, morphogenesis, and virulence, and identifies novel targets for antifungal drug development.

Chemical Genomics
Chemical genomics leverages small molecules to probe gene function and understand biological systems.
We use chemical genomics to map genetic interaction networks that control key virulence traits, and to link biologically active small molecules to their targets.
Small molecules provide a powerful approach to detect genetic interactions when a mutant allele of one gene causes an unexpected phenotype in the presence of a small molecule. Chemical genomics is crucial to mapping genetic interactions in organisms for which classical genetic approaches are hampered by the lack of a complete sexual cycle, as with C. albicans. Chemical genomics also provides a powerful strategy to identify the targets of small molecules based on the principle that reducing the dosage of a target will confer hypersensitivity to the cognate small molecule.
Our work reveals key principles by which connectivity in genetic networks can illuminate the genetic architecture of biological pathways, and identifies novel regulators of drug resistance and morphogenesis, as well as new therapeutic targets.

Mechanisms of Drug Resistance and Disease
Treatment of invasive fungal infections is notoriously difficult, with only three classes of antifungals for invasive fungal infections, compared to more than two dozen classes for bacterial infections. Resistance to each class of antifungal has emerged as a clinical problem.
We use detailed molecular genetic approaches to dissect the mechanisms that allow fungal pathogens to evade the drugs we use to kill them, and cause life-threatening infectious disease.
As just one example, our work reveals that the molecular chaperone Hsp90 is the Achilles heel of fungal pathogens, as it orchestrates the cellular circuitry required for stress response, drug resistance, morphogenesis, and virulence. Hsp90 enables drug resistance by stabilizing key regulators of cellular signaling such as the protein phosphatase calcineurin and mitogen activated protein kinase Mkc1. Hsp90 governs morphogenesis by repressing Ras1-PKA signaling and through more specialized cellular circuitry. Hsp90 function is subject to complex regulation by co-chaperones and by post-translational modification, including phosphorylation and acetylation.
We also leverage chemical biology approaches to screen for small molecules that reverse drug resistance, and to link the bioactive small molecules to their cellular target. Many small molecules with antifungal activity also modulate key virulence traits such as morphogenesis.
Our focus on fungal pathogenesis also extends to understanding the mechanisms by which fungi sense and respond to temperatures in the human host, form drug-resistant biofilms on catheters and indwelling medical devices, and interact with host immune cells.

Microbiome in Health and Disease
The human body is home to a remarkable number of microbes, collectively referred to as the human microbiome, which have a profound impact on diverse facets of human biology and disease.
We have developed powerful approaches to characterize the fungal microbiome leveraging high-throughput sequencing of the ribosomal RNA internal transcribed spacer 1 (ITS1), coupled with phenotypic and genotypic analyses fungal isolates.
This clinical genomic approach has allowed us to track fungal communities in patient populations over the course of clinical exacerbation and treatment. This reveals that fungal communities found in the lung of patients with respiratory diseases are distinct from those in healthy people. It also reveals mechanisms of pathogen adaptation over the course of chronic infection.

Structure-Guided Drug Design
Solving three-dimensional structures of drug targets allows for rational and precise drug discovery and development.
We established that targeting Hsp90 provides a powerful strategy to treat fungal infections. In species of Candida and Aspergillus, genetic depletion or pharmacological inhibition of Hsp90 abolishes resistance to the most widely deployed antifungals, including azoles, echinocandins, and polyenes. We have demonstrated that inhibiting Hsp90 function renders resistant fungal pathogens more responsive to treatment, impedes the evolution of drug resistance, and leads to clearance of otherwise lethal infections.
Hsp90 is an attractive target for drug development, and there has been phenomenal progress in the discovery of potent compounds that selectively inhibit this chaperone, driven in large part by anti-cancer activity of Hsp90 inhibitors. All Hsp90 inhibitors currently undergoing clinical development have been optimized to inhibit the human chaperone and exhibit relatively poor activity against fungi.
We have assembled an interdisciplinary team to solve the crystal structure of the N-terminal domain of C. albicans Hsp90. Guided by the key differences in the X-ray crystal structures of fungal and human Hsp90, we have been able for the first time to design, synthesize and biochemically characterize lead inhibitors of fungal Hsp90 with >10-fold selectivity relative to the human isoform.

Experimental Evolution
Experimental evolution focuses on testing hypotheses regarding evolution using controlled laboratory experiments.
Drug combinations are emerging as a promising strategy to treat fungal infections, but the mechanisms by which they evolve and the fitness consequences have remained largely enigmatic.
We have leveraged experimental evolution coupled with genome sequencing and high-resolution fitness measurements to map the genetic architecture of adaptation to antifungal drug combinations. We have complemented these studies with genome-scale analyses of how drug resistance evolves in the human host, and have identified fitness trade-offs in which the evolution of drug resistance is accompanied by hypersensitivity to killing by macrophages.
This work provides a foundation for predicting and preventing the evolution of drug resistance, and reveals fitness trade-offs that may minimize the evolution and persistence of resistance in pathogen populations.