The Segregation Problem and Plasmid Partition
The faithful segregation of a full complement of chromosomes to every cell at cell division is a fundamental problem for all organisms. We study the molecular mechanisms that segregate, or "partition", bacterial plasmids. Our system is the P1 plasmid in Escherichia coli, because it is a genetically and biochemically tractable model for the most predominant type of partition system in bacteria.
P1 Partition
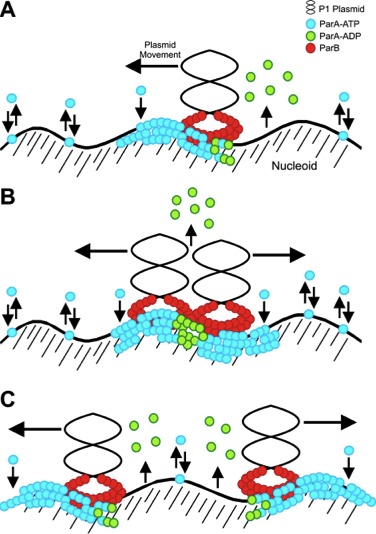
The partition system consists of two proteins, ParA and ParB, that act on a plasmid centromere-like site, called parS.
ParB binds specifically to parS, and this complex is the substrate for ParA, an ATPase, that directs plasmid movement. Our current model, based on genetics and biochemistry, is that plasmids "surf" over the surface of the bacterial nucleoid via that action of ParA (cartooned at right). P1 Par is the paradigm for studies of plasmid partition in bacteria.
Why plasmids?
We study plasmids because they play important roles in the physiology and pathogenicity of bacteria. Plasmids provide functions that are necessary for the survival of their bacterial hosts, notably genes for virulence and antibiotic resistance that can have a large impact on human health. The virulence plasmids of Yersinia pestis (bubonic plague), Salmonella typhimurium, and E. coli O157:H7, for example, require a Par system that is homologous to that of P1 for their stability. Plasmids also carry beneficial genes; for example, those of the Rhizobiaceae bacterial family, which carry genes essential for nitrogen fixation in plants (and thus human health), require P1-like partition systems for segregation. We believe all these systems will work by the same basic mechanism.
The faithful segregation of a full complement of chromosomes to every cell at cell division is a fundamental problem for all organisms. We study the molecular mechanisms that segregate, or "partition", bacterial plasmids. Our system is the P1 plasmid in Escherichia coli, because it is a genetically and biochemically tractable model for the most predominant type of partition system in bacteria.
P1 Partition
The partition system consists of two proteins, ParA and ParB, that act on a plasmid centromere-like site, called parS.
ParB binds specifically to parS, and this complex is the substrate for ParA, an ATPase, that directs plasmid movement. Our current model, based on genetics and biochemistry, is that plasmids "surf" over the surface of the bacterial nucleoid via that action of ParA (cartooned at right). P1 Par is the paradigm for studies of plasmid partition in bacteria.
Why plasmids?
We study plasmids because they play important roles in the physiology and pathogenicity of bacteria. Plasmids provide functions that are necessary for the survival of their bacterial hosts, notably genes for virulence and antibiotic resistance that can have a large impact on human health. The virulence plasmids of Yersinia pestis (bubonic plague), Salmonella typhimurium, and E. coli O157:H7, for example, require a Par system that is homologous to that of P1 for their stability. Plasmids also carry beneficial genes; for example, those of the Rhizobiaceae bacterial family, which carry genes essential for nitrogen fixation in plants (and thus human health), require P1-like partition systems for segregation. We believe all these systems will work by the same basic mechanism.
Surfing on the bacterial chromosome