Research Overview
RNAi and Small RNAs
In addition to regulating the mRNA and protein levels of target genes, emerging work in a variety of organisms indicates that small RNA-mediated activities also act in the nucleus to regulate chromatin and transcription. The modulation of chromatin by small RNAs appears to impact such key processes as gametogenesis, cell division, and cell fate specification, and is thus likely to play critical roles in both normal development and disease. Despite this importance, we have much to learn about how small RNA pathways regulate chromatin throughout animal development or about the functional consequences of such regulation. We use the tiny soil nematode C. elegans to examine how small RNA pathways regulate chromatin and impact animal development.

Discoveries on the mechanism of RNAi led to the 2006 Nobel Prize. Notably, these amazing studies were focused on embryo development, but unexpectedly revealed a previously unappreciated means of turning genes off. RNAi has revolutionized molecular biology, and highlights the importance of fundamental research.
To learn more about RNAi, click here.
Epigenetics
Interested in learning more about epigenetics? We don't blame you, it's a super cool topic! Here's a great basic primer on epigenetics that anyone can read and understand (even you, non-scientist friends!).
Worms!

You might not want to hear this, but worms are remarkably similar to humans! Worms may be small, but they are mighty research tools that help us to understand how various processes occur in humans. Click here to read a fantastic primer on using worms in the lab. For more worm resources, click here, here, and here.
Current Research
Investigating the Functions of Argonaute Proteins
Projects are available to study uncharacterized Argonautes, both at the systems level and on an AGO-by-AGO basis. We use an integrated combination of classical genetics, genetic engineering [Cas-CRISPR, transgene studies], microscopy, molecular biology/biochemistry [IP, AP/MS, in vitro assays], and genomics [RIP-seq, CLIP-seq, mRNA-seq, ChIP-seq, and more] to study these fascinating proteins.

Argonaute proteins are the hub of amazing ribonucleoprotein (RNP) complexes with diverse functions. To learn more about AGOs in the worm, check out this review by JC and Dr. Elaine Youngman.
AGO projects are funded by:
The Argonaute CSR-1: An Anti-Silencer
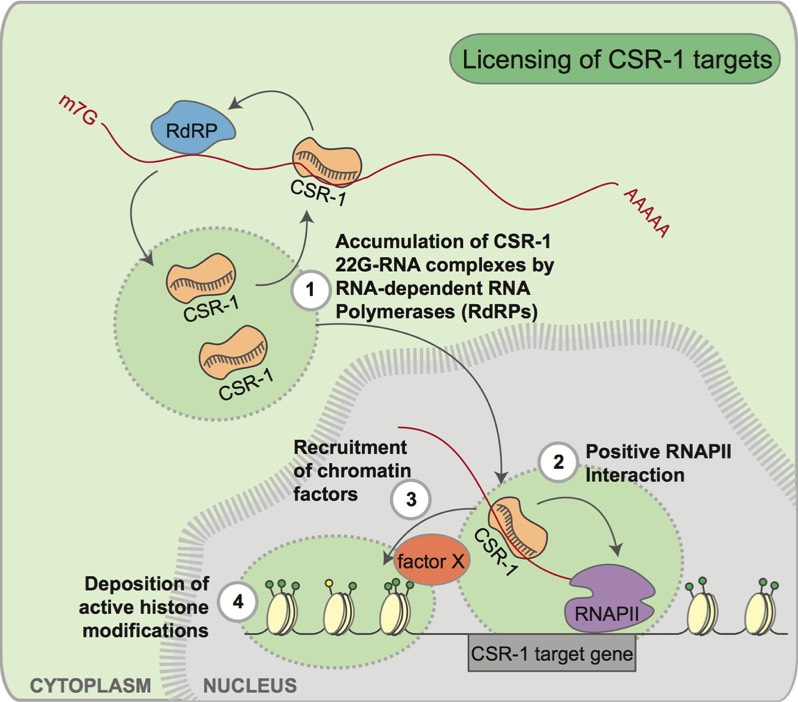
Much to our surprise, our earlier studies indicated that CSR-1 does not silence the expression of its targets, but instead promotes their expression [i.e., CSR-1 is an Anti-Silencer!]. We determined this using an in vivo tethering assay, where we recruited CSR-1 to one specific gene in the genome. When CSR-1 was not recruited to this gene, it would normally be silenced by the piRNA pathway. The piRNA pathway can be thought of as the adaptive immune system of the germline, surveying for invading nucleic acid sequences, and stopping them in their tracks before they damage the genome. Remarkably, when CSR-1 was tethered to the gene, it was protected from silencing by the piRNA pathway and remained expressed in the germline! This observation shifts paradigms: Argonautes are not only capable of silencing gene expression, but can also promote the expression of their target genes! We are now also examining the role of CSR-1 in other nematodes using comparative genomics, and have found evidence for a conserved role for CSR-1 in promoting gene expression. Currently, we are dissecting the molecular mechanisms by which CSR-1 impacts chromatin and gene expression, and have identified new nuclear factors that act in the CSR-1 pathway, such as the conserved Intron Binding Protein/Helicase EMB-4/Aquarius.
Projects are available to study the mechanisms of CSR-1 activity on chromatin and gene expression. We are using forward and reverse genetics as well as biochemical approaches (co-IP, AP/MS) to identify factors that function in the CSR-1 pathway. Genetic engineering [Cas-CRISPR, transgene studies] of CSR-1 and in vitro assays enable us to perform structure-function analyses of CSR-1. Microscopy and genomics [RIP-seq, CLIP-seq, mRNA-seq, ChIP-seq] allow us to determine how CSR-1 impacts chromatin and the transcription of its germline targets. Other projects are available to explore the piRNA pathway, especially regarding the adaptive steps of the process. We also have evolutionary projects, using comparative genomics to understand how the CSR-1 and piRNA pathways function to defend the germline genome.
CSR-1 is an extremely interesting AGO protein. To learn more about CSR-1, check out this review by Chris, Monica and JC. To learn more about how CSR-1 antagonizes the piRNA pathway to promote germline gene expression, check out this commentary by Chris, Monica and JC.
CSR-1 projects are funded by:


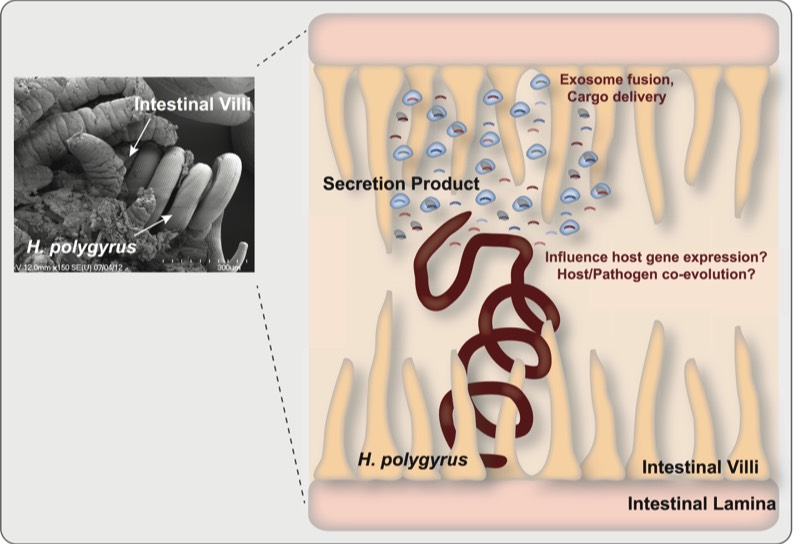
Communication through Extracellular small RNAs
H. polygyrus uses small RNAs to communicate with its host (mice) and manipulate its environment (the intestine) in favor of its persistence in the host. To learn more about H. polygyrus, click here.
H. polygyrus projects are funded by:

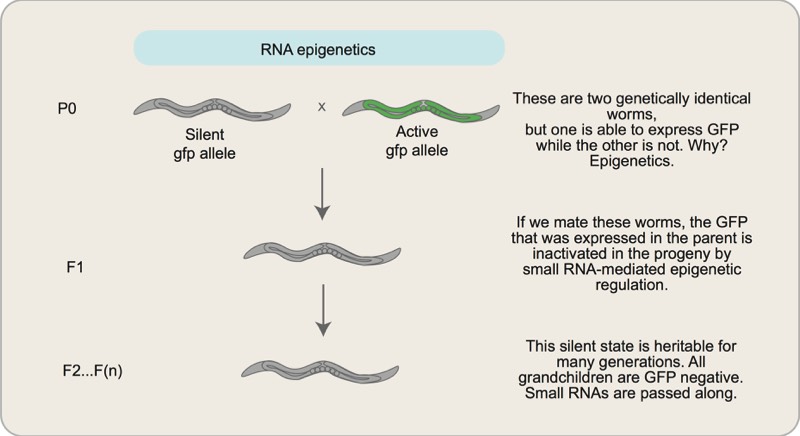
Small RNAs, Environmental Inputs, and Epigenetic Inheritance

What stresses do you think Wormzilla is experiencing here?