Diagnosis - RNA based Prion Disease Diagnostic Tools
Section Navigator> Diagnosis>
> Endogenous RNA extract & PMAC
> mRNA markers
> RNA aptamers
RNA Aptamers
RNA aptamers are strands of RNA that have been evolved (in vitro) in a process known as SELEX (systematic evolution of ligands by exponential enrichment) to be able to bind specifically to any biological compound with high affinity (1).
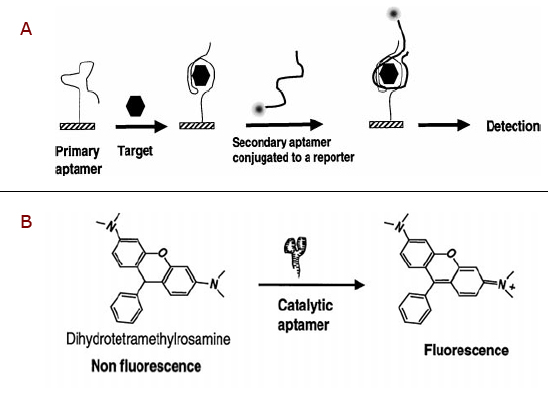
Figure 1. Two models in which aptamers can be used to identify a target and elicit a reporter signal as means of detecting the target (Adapted from Jayasena (2)). In A) a primary aptamer bound to a solid support recognizes the target molecule, then a secondary aptamer with reporter capabilities binds to the primary aptamer-target complex to yield a detection signal of the amount of target molecules present. B) An altertative approach in which a catalytic aptamer is used to convert a non-fluorescence molecule into a fluorescent one, only when bound to the target.
Aptamers rival the technology of antibodies but also has the ability to undergo chemical modifications and can also bind to proteins and ligands that normally would have been protected by the host immune system such as PrPSC, something antibodies have difficulties accomplishing (3). The high binding affinity of RNA aptamers to the mutant and natural forms of Prion proteins (3) and their abilities to be chemically modified makes them ideal candidates as diagnostic tools of Prion disease. Researchers have been able to develop an antibody capable of recognizing between the normal and the mutant Prion protein (3). Current diagnostics for Prion disease requires the use of recently developed antibodies, which require denaturing agents on the sample and can only detect the protein kinase resistant form of the mutant Prion protein (3). The advantage that RNA aptamers may hold over current diagnostics is the evidence that RNA aptamers can retain high binding affinities without the use of denaturing reagents, which can alter results (3). Moreover because RNA aptamers can be evolved to have strong binding to almost any compound, RNA aptamers can detect more than just the protein kinase resistant form of Prion proteins that antibodies are restricted to detect (3). Furthermore, because RNA aptamers are synthesized in vitro, this allows them to be subject to chemical modifications (1). Modifications include probes and detectors such as biotinylation (4). Complexes with the biotin domain have the ability to bind to solid supports, allowing for their detection and their quantification through use of conventional western blotting (4). RNA aptamers possess the ability to bind and identify mutant Prion proteins, as well the ability to be detected and quantified through convention western blotting and the ability to overcome barriers encountered by current detection methods. Thus RNA aptamers are a promising solution for Prion protein detection.
References:
(1)Bunka DHJ, Stockley PG. Aptamers come of age – at last. Nature Reviews Microbiology 2006; 4:588-596. (2)Jayasena, SD. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clinical Chemistry 1999; 45:1628-1650.(3)Sayer NM, Cubin M, Rhie A, Bullock M, Tahiri-Alaoui A, James W. Structural determinants of conformationally selective, Prion-binding aptamers. The Journal of Biological Chemistry 2004; 279:13102-13109. (4)Sekiya S, Noda K, Nishikawa F, Yokoyama T, Kumar PKR, Nishikawa S. Characterization and application of a novel RNA aptamer against the mouse Prion protein. J. Biochem. 2006; 139:383-390.

