Reviews of current RNA based technologies used in Prion Disease Research
Section Navigator> Scientific Reviews>
> Endogenous ssRNA and shsRNA
> mRNA markers in Prion Diease
> RNA aptamers and Prion Diesase
> RNA interferance and Prion Disease
RNA Aptamers in Prion Disease
Prion disease is an infectious, incurable, neurodegenerative disease that affects and can be transmitted between humans and other mammals (1). Prion diseases are a class of neurological diseases characterized by their viral-like activity, however its mechanism is not viral in nature but rather believed to be based on a mutant protein (PrPSC) that is able to convert normal Prion protein (PrPC) (function unknown) to fold into the mutant form despite that their amino acid sequences maybe identical (1). This mutant is believed to aggregate and cause damage to nearby tissue (2) but the exact mechanism is not completely understood. There exists no treatment or ability to effectively diagnose the disease before signs of neurodegeneration or transmission has occurred (3). RNA aptamers are strands of RNA that have been evolved (in vitro) in a process known as SELEX (systematic evolution of ligands by exponential enrichment) to be able to bind specifically to any biological compound with high affinity (4).
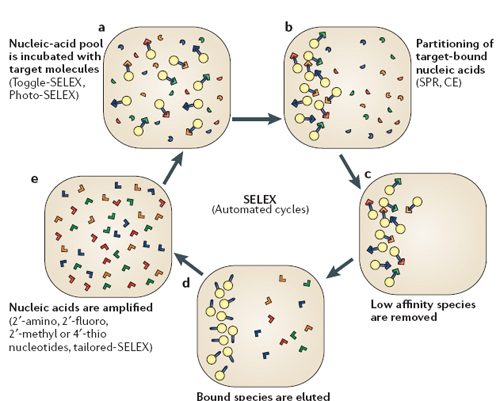
Figure 1. Overview of the SELEX process, a library of random sequence RNA strands are incubated with the target molecule. Interactions occur between RNA and target, some maybe strong. Weak affinity RNA ligands are washed off. The remaining RNA strands are eluted and amplified. During amplification modifications can be induced such as 2’-amino groups to protect against degradation, as well errors during amplification can be induced to lead to diversity of the ligand library. This process is completed until a family of strong binding ligands are achieved (Adapted from Bunka et al. (4)).
Aptamers rival the technology of antibodies but also has the ability to undergo chemical modifications and can also bind to proteins and ligands that normally would have been protected by the host immune system such as PrPSC, something antibodies have difficulties accomplishing (3). In understanding the types of RNA aptamers that bind to PrPSC we can tailor RNA aptamers that are sensitive enough to act as reliable diagnostic tools (3) for screening PrPSC and to act as molecules that can target and eliminate PrPSC (5). Furthermore, the study of aptamers in understanding what properties are needed to bind to the Prion protein may elucidate physiological roles of the prion protein (6).
Aptamers can distinguish between PrPC and PrPSC
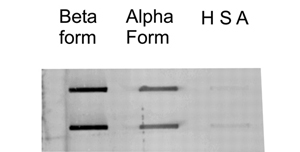
Figure 2. Slot blot assay of RNA aptamer SAF-93 exhibiting preferential binding to the beta-PrP form (mutant PrP) over the alpha-PrP form (normal PrP). Human serum albumin (HSA) used as a control in the assay. (Adapted from Sayer et. al.(3)).
PrPC and PrPSC differ in only how they are folded, PrPC consists mainly of a-helical secondary structures while PrPSC differs by having ß-sheet structures (1), in order to distinguish between the two, a molecule with a very high binding affinity for one over the other is required. Rhie et al. (7) developed an aptamer (SAF-93) that binds to PrPSC ten times greater than it binds to PrPC. SAF-93 is a 2'-fluoro-modified RNA aptamer that was evolved in vitro from a randomized library of RNA sequences through SELEX against scrapie-associated fibrils (SAF) (7).
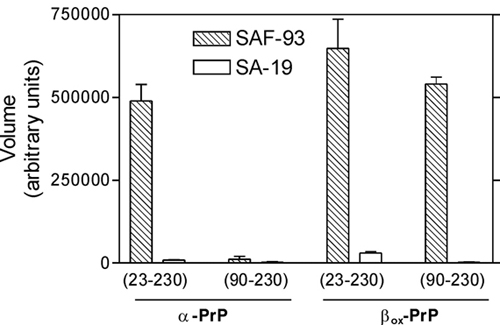
Figure 3. Slot blot analysis of RNA aptamer SAF-93 binding to various truncated parts of both versions of the prion protein. Values were determined from intensity of the bands using ImageQuant software. This figure shows that SAF-93 preferably binds to the 90-230 region of the beta-PrP(misfolded), and does not bind to the same region of the alpha-PrP(properly folded)(Adapted from Rhie et. al.(7)).
SAF-93 is found to bind to two sites on PrPSC, the first is found on residues 90-110 in which a structure is exposed in PrPSC but inaccessible in PrPC, and the second binding site consists of ß-sheets located on the C-terminus of residue 110 which occurs only in PrPSC(7). The ability of aptamers to distinguish between the two proteins makes it an ideal candidate for diagnostic and therapeutic roles.
Aptamer Stability and Modifications
Being RNA molecules, RNA aptamers can be subject to digestion, degradation or renal clearance in vivo, an issue when considering aptamers as potential therapeutic or diagnostic tools (4). Fortunately because aptamers are raised in vitro unlike antibodies, they can be chemically modified. 2'-amino, 2'-o-methyl, 4'-thio, and 2'-fluoro-modifications on pyrimidines of RNA aptamers are known to increase stability of these molecules in the body (4)(8), this allows aptamers the ability to have a controlled half-life. Furthermore the use of Spiegelmers (L-ribose rather than D-ribose) nucleic acids can avoid digestion by enzymes and further biostability (4). Aptamers can also be biotinylated, the process of adding biotin to the molecule which can bond strongly to a solid support and used to detect PrPC or PrPSC in the tissue (8). These modifications give aptamers the ability to be used as in vivo diagnostic and therapeutic tools.
RNA Aptamers as diagnostics for Prion Disease
The high binding affinity of RNA aptamers to the mutant and natural forms of prion proteins and their abilities to be chemically modified makes them ideal candidates as diagnostic tools of Prion disease. Sekiya et al. (8) demonstrates that their biotinylated aptamer (60-3Fb) and a 32P-label 60-3F aptamer were both able to detect PrPC through conventional western blotting similar to antibodies. The authors note that because aptamers and their chemical modifications can be synthesized on a larger scale than antibodies, their role as detectors are more useful than antibodies (8). However, this approach cannot identify the presence or absence of PrPSC. The 60-30Fb and 32P-label 60-3F aptamers may be useful in clinical settings, and has applications in research involving the need to detect the PrPC protein. But it is not a sufficient detection tool for prion disease.
Sayer et al. has developed a version of the SAF-93 aptamer (which preferentially binds to PrPSC ) labeled as SAF-93(1-34,35bioU,36-60) to incorporate biotinylation modifications and shortened enough to be able to chemically synthesize the molecule without losing its binding ability (3). SAF-93(1-34,35bioU,36-60) offers advantages to recently developed anti-PrP and anti-PrPSC antibodies which rely on denaturing agents and the protein kinase resistant form of PrPSC to function(3). Since SAF-93(1-34,35bioU,36-60) and other aptamers are able to bind to PrPSC that does not rely on its protein kinase (PK) resistance, these aptamers can recognize non-PK-resistant forms of PrPSC (3). Furthermore because SAF-93(1-34,35bioU,36-60) can bind to PrPSC in natural conditions (without denaturing reagents), this aptamer offers the possibility to bind to PrPSC in vivo, a possibility that has yet to be rivaled but has yet to be explored.
RNA Aptamers as therapeutic tools for Prion Disease

Figure 4. Result of a plot of PrPres generated from cell-free conversion assay in the presence or absence of RNA aptamer SAF-93. Graph shows that SAF-93 is able to significantly reduce PrPC to PrPres conversion. (Adapted from Rhie et. al.(7)).
Already Rhie et al. and Proske et al. have shown evidence to support the abilities of RNA aptamers to inhibit PrPC to PrPSC conversion and reduce PrPSC formation respectively (7)(5). RNA aptamers hold great potential as therapeutic drug: they have the ability to discriminate between the two forms of PrP (7), their RNA backbone may not trigger immune responses (4), their easy clearance and degradation can reduce the possibility of drug accumulation (4), their ability to be chemically modified (8), and their ability to be chemically synthesized in bulk (8) makes RNA aptamers one of the best candidates for drug therapy.
Aptamers in understanding PrPC role
Mercey et al. used of SELEX to generate high binding affinity RNA aptamers of Kd 20nM sensitivity to elucidate the minimum requirements for ligand binding (6). They found that due to the fast dissociation and association rates, it is quite possible that their aptamer (RM312) is simulating interactions of a endogenous nucleic acid ligand that PrPC normally interacts with (6). This suggests that PrPC might have a role in interacting with nucleic acids. However, RNA aptamers are regarded to be able to bind to almost any biological compound, if this is the case with PrPC then a nucleic acid ligand that naturally interacts with PrPC (a membrane bound protein) (1) may not exist.
Therefore, RNA aptamers offer the ability to distinguish between the Prion proteins, and to be modified to become diagnostic and therapeutic tools. RNA aptamers hold an advantage over antibody technology by not being restricted by immunoprotected proteins, by having higher binding affinities, and by being easier modify and produce chemically. This illustrates the advantages that RNA aptamers provide over antibody technologies in investigating Prion diseases, and also illustrates the abilities of RNA aptamers to succeed as diagnostic and therapeutic options.
References
1. Watts JC, Balachandran A, Westaway D. The Expanding Universe of Prion Diseases. PloS Pathogens 2006; 2:152-163.
2. Alberts B, Johnson A, Lewis J, Raff M, Roberts K, and Walter P. Molecular Biology of the Cell. 4th ed. New York: Garland Publishing; 2002. pp 362-363.
3. Sayer NM, Cubin M, Rhie A, Bullock M, Tahiri-Alaoui A, James W. Structural determinants of conformationally selective, prion-binding aptamers. The Journal of Biological Chemistry 2004; 279:13102-13109.
4. Bunka DHJ, Stockley PG. Aptamers come of age – at last. Nature Reviews Microbiology 2006; 4:588-596. 5. Proske D, Gilch S, Wopfner F, Schatzl HM, Winnacker EL, Famulok M. Prion-protein-specific aptamer reduces PrPsc formation. ChemBioChem 2002; 3:717-725.
6. Mercey, R. Lantier I, Maurel MC, Grosclaude J, Lantier F, Marc D. Fast, reversible interaction of prion protein with RNA aptamers containing specific sequence patterns. Archives of Virology 2006; 151:2197-2214.
7. Rhie A, Kirby L, Sayer N, Wellesley R, Disterer P, Sylvester I, Gill A, Hope J, James W, Tahiri-Alaoui A. Characterization of 2'-flouro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. The Journal of Biological Chemistry 2003; 278:39697-39705.
8. Sekiya S, Noda K, Nishikawa F, Yokoyama T, Kumar PKR, Nishikawa S. Characterization and application of a novel RNA aptamer against the mouse prion protein. J. Biochem. 2006; 139:383-390.

